This page was generated from us_coasts.ipynb.
Interactive online version:
CONUS Coasts#
US States and Coastlines#
Census TIGER (Topologically Integrated Geographic Encoding and Referencing database)
[1]:
from __future__ import annotations
import warnings
from pathlib import Path
import geopandas as gpd
warnings.filterwarnings("ignore", message=".*initial implementation of Parquet.*")
root = Path("input_data")
root.mkdir(exist_ok=True)
BASE_PLOT = {"facecolor": "k", "edgecolor": "b", "alpha": 0.2, "figsize": (12, 6)}
CRS = "epsg:5070"
def tiger_shp_2022(data):
url = f"https://www2.census.gov/geo/tiger/TIGER2022/{data.upper()}/tl_2022_us_{data}.zip"
return gpd.read_file(url)
cfile = Path(root, "us_state.feather")
if cfile.exists():
state = gpd.read_feather(cfile)
else:
state = tiger_shp_2022("state")
state.to_feather(cfile)
[2]:
ax = state.to_crs(CRS).plot(**BASE_PLOT)
ax.axis("off")
ax.margins(0)
[3]:
cfile = Path(root, "us_coastline.feather")
if cfile.exists():
coastline = gpd.read_feather(cfile)
else:
coastline = tiger_shp_2022("coastline")
coastline.to_feather(cfile)
[4]:
ax = coastline.to_crs(CRS).plot(figsize=(12, 6))
ax.axis("off")
ax.margins(0)
Clip to CONUS#
[5]:
from shapely.geometry import box
conus_bounds = box(-125, 24, -65, 50)
cfile = Path(root, "conus_states.feather")
if cfile.exists():
conus_states = gpd.read_feather(cfile)
else:
conus_states = state[state.within(conus_bounds)]
conus_states.to_feather(cfile)
[6]:
ax = conus_states.to_crs(CRS).plot(**BASE_PLOT)
ax.axis("off")
ax.margins(0)
[7]:
conus_coastline = coastline[coastline.within(conus_bounds)]
cfile = Path(root, "us_coast_states.feather")
if cfile.exists():
coast_states = gpd.read_feather(cfile)
else:
coast_states = state[state.intersects(conus_coastline.union_all)]
coast_states.to_feather(cfile)
[8]:
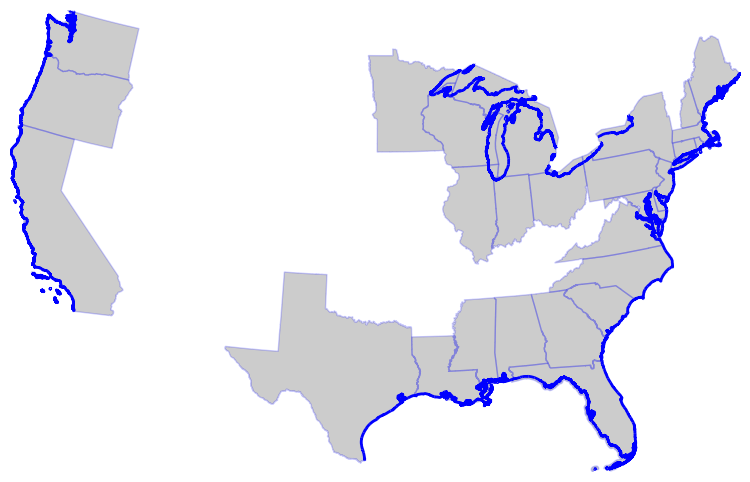
ax = coast_states.to_crs(CRS).plot(**BASE_PLOT)
conus_coastline.to_crs(CRS).plot(ax=ax, edgecolor="b", lw=2, zorder=1)
ax.axis("off")
ax.margins(0)
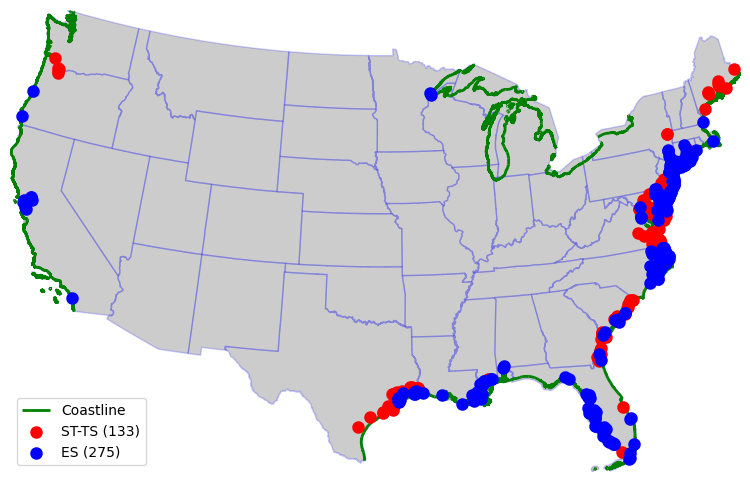
Tidal and Estuary USGS stations#

We need to look at the Water Services API.
[9]:
import pandas as pd
from pygeohydro import NWIS
from pygeohydro.exceptions import ZeroMatchedError
cfile = Path(root, "coast_stations.feather")
if cfile.exists():
coast_stations = gpd.read_feather(cfile)
else:
queries = [
{
"stateCd": s.lower(),
"siteType": "ST-TS,ES",
"hasDataTypeCd": "dv",
"outputDataTypeCd": "dv",
}
for s in coast_states.STUSPS
]
nwis = NWIS()
def get_info(q):
try:
return nwis.get_info(q, True)
except ZeroMatchedError:
return None
sites = pd.concat(get_info(q) for q in queries if q is not None)
coast_stations = gpd.GeoDataFrame(
sites,
geometry=gpd.points_from_xy(sites.dec_long_va, sites.dec_lat_va),
crs="epsg:4269",
)
coast_stations.to_feather(cfile)
st = coast_stations[["site_no", "site_tp_cd", "geometry"]].to_crs(CRS)
ts = st[st.site_tp_cd == "ST-TS"].drop_duplicates()
es = st[st.site_tp_cd == "ES"].drop_duplicates()
[10]:
ax = conus_states.to_crs(CRS).plot(**BASE_PLOT)
coastline[coastline.within(conus_bounds)].to_crs(CRS).plot(ax=ax, edgecolor="g", lw=2, zorder=1)
ts.plot(ax=ax, lw=3, c="r")
es.plot(ax=ax, lw=3, c="b")
ax.legend(["Coastline", f"ST-TS ({ts.shape[0]})", f"ES ({es.shape[0]})"], loc="best")
ax.axis("off")
ax.margins(0)
ax.figure.savefig(Path("_static", "us_coasts.png"), dpi=300, bbox_inches="tight", facecolor="w")
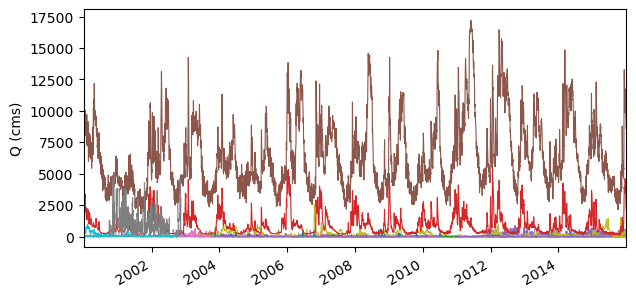
Mean daily discharge for all stations#
[11]:
import numpy as np
import pandas as pd
cfile = Path(root, "discharge.parquet")
dates = ("2000-01-01", "2015-12-31")
if cfile.exists():
discharge = pd.read_parquet(cfile)
else:
nwis = NWIS()
discharge = nwis.get_streamflow(
coast_stations.site_no,
dates,
)
discharge[discharge < 0] = np.nan
discharge.to_parquet(cfile)
[12]:
ax = discharge.plot(legend=False, lw=0.8, figsize=(7, 3.5))
ax.set_ylabel("Q (cms)")
ax.set_xlabel("")
ax.margins(x=0)
Let’s find the station that has the largest peak value and see it on a map.
[13]:
station_id = discharge.max().idxmax().split("-")[1]
coast_stations[coast_stations.site_no == station_id].iloc[0]["station_nm"]
[13]:
'COLUMBIA RIVER AT PORT WESTWARD, NEAR QUINCY, OR'
[14]:
import pygeohydro as gh
lat, lon = coast_stations[coast_stations.site_no == station_id].iloc[0][
["dec_lat_va", "dec_long_va"]
]
bbox = (lon - 0.2, lat - 0.2, lon + 0.2, lat + 0.2)
nwis_kwds = {"hasDataTypeCd": "dv", "outputDataTypeCd": "dv", "parameterCd": "00060"}
station_map = gh.interactive_map(bbox, nwis_kwds=nwis_kwds)
[15]:
station_map
[15]:
Make this Notebook Trusted to load map: File -> Trust Notebook
River network data#

Basin#
[16]:
import pynhd as nhd
nldi = nhd.NLDI()
cfile = Path(root, "basin.feather")
if cfile.exists():
basin = gpd.read_feather(cfile)
else:
basin = nldi.get_basins(station_id)
basin.to_feather(cfile)
[17]:
ax = basin.to_crs(CRS).plot(**BASE_PLOT)
ax.axis("off")
ax.margins(0)
[18]:
import folium
folium.GeoJson(basin.geometry).add_to(station_map)
station_map
[18]:
Make this Notebook Trusted to load map: File -> Trust Notebook
Main river network#
[19]:
cfile = Path(root, "flowline_main.feather")
if cfile.exists():
flw_main = gpd.read_feather(cfile)
else:
flw_main = nldi.navigate_byid(
fsource="nwissite",
fid=f"USGS-{station_id}",
navigation="upstreamMain",
source="flowlines",
distance=4000,
)
flw_main.to_feather(cfile)
Tributaries#
[20]:
cfile = Path(root, "flowline_trib.feather")
if cfile.exists():
flw_trib = gpd.read_feather(cfile)
else:
flw_trib = nldi.navigate_byid(
fsource="nwissite",
fid=f"USGS-{station_id}",
navigation="upstreamTributaries",
source="flowlines",
distance=4000,
)
flw_trib.to_feather(cfile)
flw_trib["nhdplus_comid"] = flw_trib["nhdplus_comid"].astype("float").astype("Int64")
[21]:
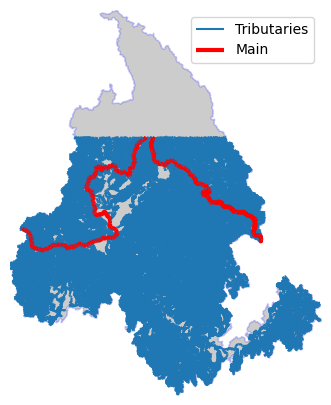
BASE_PLOT["figsize"] = (8, 5)
ax = basin.plot(**BASE_PLOT)
flw_trib.plot(ax=ax)
flw_main.plot(ax=ax, lw=3, color="r")
ax.legend(["Tributaries", "Main"])
ax.axis("off")
ax.margins(0)
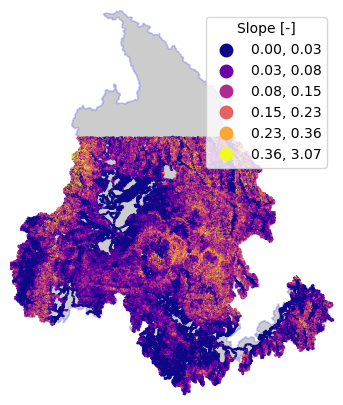
NHDPlus Value Added Attributes (VAA)#
[22]:
cfile = Path(root, "nhdplus_vaa.parquet")
nhdplus_vaa = nhd.nhdplus_vaa(cfile)
flw_trib = flw_trib.merge(nhdplus_vaa, left_on="nhdplus_comid", right_on="comid", how="left")
flw_trib.loc[flw_trib.slope < 0, "slope"] = np.nan
[23]:
ax = basin.plot(**BASE_PLOT)
flw_trib.plot(
ax=ax,
column="slope",
scheme="natural_breaks",
k=6,
cmap="plasma",
legend=True,
legend_kwds={"title": "Slope [-]"},
)
ax.axis("off")
ax.margins(0)