This page was generated from nldas.ipynb.
Interactive online version:
NLDAS2 Forcing#
[1]:
from __future__ import annotations
from pathlib import Path
import pynldas2 as nldas
from pygeohydro import WBD
The NLDAS2 database provides forcing data at 1/8th-degree grid spacing and range from 01 Jan 1979 to present. Let’s take a look at NLDAS2 grid mask that includes land, water, soil, and vegetation masks:
[2]:
grid = nldas.get_grid_mask()
grid
[2]:
<xarray.Dataset> Size: 2MB
Dimensions: (lon: 464, lat: 224, time: 1, bnds: 2)
Coordinates:
* lon (lon) float32 2kB -124.9 -124.8 -124.7 ... -67.31 -67.19 -67.06
* lat (lat) float32 896B 25.06 25.19 25.31 ... 52.69 52.81 52.94
* time (time) datetime64[ns] 8B 2000-01-01
spatial_ref int64 8B 0
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) datetime64[ns] 16B ...
NLDAS_mask (time, lat, lon) float32 416kB ...
CONUS_mask (time, lat, lon) float32 416kB ...
NLDAS_veg (time, lat, lon) float32 416kB ...
NLDAS_soil (time, lat, lon) float32 416kB ...
Attributes: (12/13)
missing_value: -9999.0
time_definition: constant
title: NLDAS masks and predominant vegetation/soil
institution: NASA GSFC
history: created on date: Fri Mar 8 15:58:50 2019
references: Mitchell_etal_JGR_2004; Xia_etal_JGR_2012
... ...
website: https://ldas.gsfc.nasa.gov/nldas/
MAP_PROJECTION: EQUIDISTANT CYLINDRICAL
SOUTH_WEST_CORNER_LAT: 25.0625
SOUTH_WEST_CORNER_LON: -124.9375
DX: 0.125
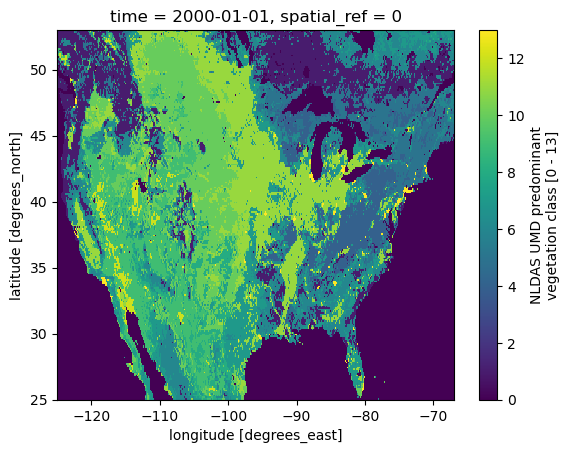
DY: 0.125For example, let’s plot the vegetation mask.
[3]:
ax = grid.NLDAS_veg.plot()
ax.figure.savefig(Path("_static", "nldas_grid.png"), facecolor="w", bbox_inches="tight")
Next, we use PyGeoHydro to get the geometry of a HUC8 with ID of 1306003:
[4]:
huc8 = WBD("huc8")
geometry = huc8.byids("huc8", "13060003").geometry.iloc[0]
PyNLDAS2 allows us to get the data for a list of coordinates using pynldas2.get_bycoords or for a region as gridded data using pynldas2.get_bygeom. Here, we use the latter. Note that if we don’t pass any variables, all variables will be downloaded. Additionally, we can pass snow=True to separate snow portion of precipitation using temperature. PyNLDAS2 provides access to NLDAS2 data from two sources: grib and netcdf. They are mostly
similar so if for some reason one of the sources is not temporarly available, you can use the other one. The default source is grib.
[5]:
clm = nldas.get_bygeom(geometry, "2010-01-08", "2010-01-08", 4326, variables="prcp", snow=True)
[6]:
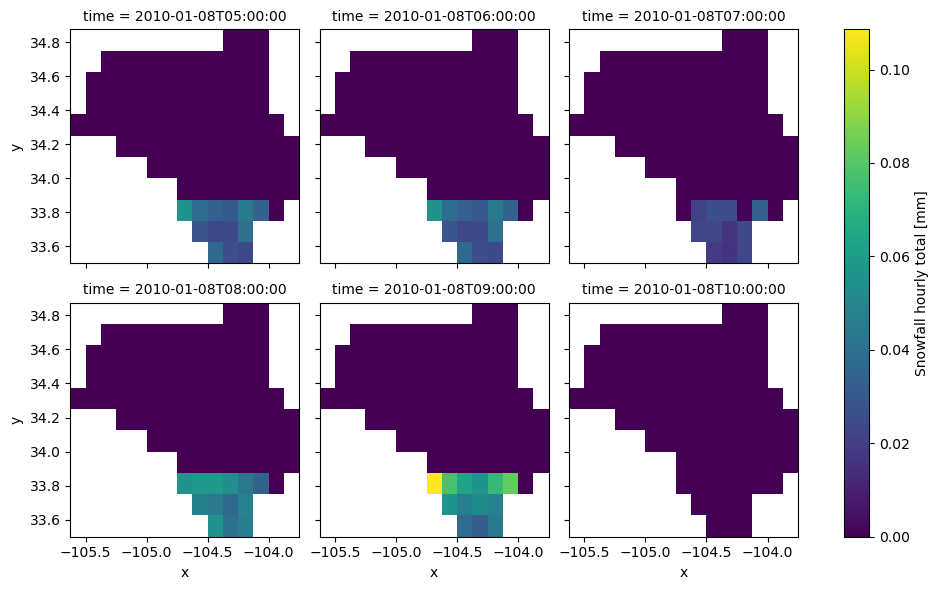
ax = clm.snow.sel(time=slice("2010-01-08T05:00:00", "2010-01-08T010:00:00")).plot(
col="time", col_wrap=3
)
ax.fig.savefig(Path("_static", "nldas_snow.png"), facecolor="w", bbox_inches="tight")