PyDaymet: Daily climate data through Daymet#






Warning
Since the release of Daymet v4 R1 on November 2022, the URL of Daymet’s server has been changed. Therefore, only PyDaymet v0.13.7+ is going to work, and previous versions will not work anymore.
Features#
PyDaymet is a part of HyRiver software stack that
is designed to aid in hydroclimate analysis through web services. This package provides
access to climate data from
Daymet V4 R1 database using NetCDF
Subset Service (NCSS). Both single pixel (using get_bycoords function) and gridded data (using
get_bygeom) are supported which are returned as
pandas.DataFrame and xarray.Dataset, respectively. Climate data is available for North
America, Hawaii from 1980, and Puerto Rico from 1950 at three time scales: daily, monthly,
and annual. Additionally, PyDaymet can compute Potential EvapoTranspiration (PET)
using three methods: penman_monteith, priestley_taylor, and hargreaves_samani for
both single pixel and gridded data.
For PET computations, PyDaymet accepts four additional user-defined parameters:
penman_monteith:soil_heat_flux,albedo,alpha,and
arid_correction.
priestley_taylor:soil_heat_flux,albedo, andarid_correction.hargreaves_samani: None.
Default values for the parameters are: soil_heat_flux = 0, albedo = 0.23,
alpha = 1.26, and arid_correction = False.
An important parameter for priestley_taylor and penman_monteith methods
is arid_correction which is used to correct the actual vapor pressure
for arid regions. Since relative humidity is not provided by Daymet, the actual
vapor pressure is computed assuming that the dew point temperature is equal to
the minimum temperature. However, for arid regions, FAO 56 suggests subtracting
minimum temperature by 2-3 °C to account for the fact that in arid regions,
the air might not be saturated when its temperature is at its minimum. For such
areas, you can pass {"arid_correction": True, ...} to subtract 2 °C from the
minimum temperature for computing the actual vapor pressure.
Both get_bygeom and get_bycoords functions save the intermediate files
returned by the web service in a local cache folder (./cache in the current
directory). The cache folder is created automatically when the functions are
called for the first time. The cache folder is used to store the intermediate
files to avoid re-downloading them. These two functions allow modifying the
web service calls via two options:
conn_timeout: Sets the connection timeout in seconds. The default value is 5 minutes. This can be increaseed for larger requests. If running these functions fails with a connection timeout error, try increasing this value.validate_filesize: IfTrue, the functions compares the file size of the previously cached files in the./cachefolder, if they exist, with their size on the remote server. If the sizes do not match, the cached files are removed and they will be re-download. By default this is set toFalsesince the files on the server rarely change. So, if a request has already been cached there shouldn’t be a need for re-donwloading them from scratch. However, if you suspect that the files on the server have changed or the functions fails to process the cached files, you can set this toTrueor manually delete the cached files in the./cachefolder.
You can find some example notebooks here. You can also try using PyDaymet without installing it on your system by clicking on the binder badge. A Jupyter Lab instance with the HyRiver stack pre-installed will be launched in your web browser, and you can start coding!
Moreover, requests for additional functionalities can be submitted via issue tracker.
Citation#
If you use any of HyRiver packages in your research, we appreciate citations:
@article{Chegini_2021,
author = {Chegini, Taher and Li, Hong-Yi and Leung, L. Ruby},
doi = {10.21105/joss.03175},
journal = {Journal of Open Source Software},
month = {10},
number = {66},
pages = {1--3},
title = {{HyRiver: Hydroclimate Data Retriever}},
volume = {6},
year = {2021}
}
Installation#
You can install PyDaymet using pip after installing libgdal on your system
(for example, in Ubuntu run sudo apt install libgdal-dev):
$ pip install pydaymet
Alternatively, PyDaymet can be installed from the conda-forge repository
using Conda:
$ conda install -c conda-forge pydaymet
Quick start#
You can use PyDaymet using command-line or as a Python library. The commanda-line provides access to two functionality:
Getting gridded climate data: You must create a
geopandas.GeoDataFramethat contains the geometries of the target locations. This dataframe must have four columns:id,start,end,geometry. Theidcolumn is used as filenames for saving the obtained climate data to a NetCDF (.nc) file. Thestartandendcolumns are starting and ending dates of the target period. Then, you must save the dataframe as a shapefile (.shp) or geopackage (.gpkg) with CRS attribute.Getting single pixel climate data: You must create a CSV file that contains coordinates of the target locations. This file must have at four columns:
id,start,end,lon, andlat. Theidcolumn is used as filenames for saving the obtained climate data to a CSV (.csv) file. Thestartandendcolumns are the same as thegeometrycommand. Thelonandlatcolumns are the longitude and latitude coordinates of the target locations.
$ pydaymet -h
Usage: pydaymet [OPTIONS] COMMAND [ARGS]...
Command-line interface for PyDaymet.
Options:
-h, --help Show this message and exit.
Commands:
coords Retrieve climate data for a list of coordinates.
geometry Retrieve climate data for a dataframe of geometries.
The coords sub-command is as follows:
$ pydaymet coords -h
Usage: pydaymet coords [OPTIONS] FPATH
Retrieve climate data for a list of coordinates.
FPATH: Path to a csv file with four columns:
- ``id``: Feature identifiers that daymet uses as the output netcdf filenames.
- ``start``: Start time.
- ``end``: End time.
- ``lon``: Longitude of the points of interest.
- ``lat``: Latitude of the points of interest.
- ``time_scale``: (optional) Time scale, either ``daily`` (default), ``monthly`` or ``annual``.
- ``pet``: (optional) Method to compute PET. Supported methods are:
``penman_monteith``, ``hargreaves_samani``, ``priestley_taylor``, and ``none`` (default).
- ``snow``: (optional) Separate snowfall from precipitation, default is ``False``.
Examples:
$ cat coords.csv
id,lon,lat,start,end,pet
california,-122.2493328,37.8122894,2012-01-01,2014-12-31,hargreaves_samani
$ pydaymet coords coords.csv -v prcp -v tmin
Options:
-v, --variables TEXT Target variables. You can pass this flag multiple
times for multiple variables.
-s, --save_dir PATH Path to a directory to save the requested files.
Extension for the outputs is .nc for geometry and .csv
for coords.
--disable_ssl Pass to disable SSL certification verification.
-h, --help Show this message and exit.
And, the geometry sub-command is as follows:
$ pydaymet geometry -h
Usage: pydaymet geometry [OPTIONS] FPATH
Retrieve climate data for a dataframe of geometries.
FPATH: Path to a shapefile (.shp) or geopackage (.gpkg) file.
This file must have four columns and contain a ``crs`` attribute:
- ``id``: Feature identifiers that daymet uses as the output netcdf filenames.
- ``start``: Start time.
- ``end``: End time.
- ``geometry``: Target geometries.
- ``time_scale``: (optional) Time scale, either ``daily`` (default), ``monthly`` or ``annual``.
- ``pet``: (optional) Method to compute PET. Supported methods are:
``penman_monteith``, ``hargreaves_samani``, ``priestley_taylor``, and ``none`` (default).
- ``snow``: (optional) Separate snowfall from precipitation, default is ``False``.
Examples:
$ pydaymet geometry geo.gpkg -v prcp -v tmin
Options:
-v, --variables TEXT Target variables. You can pass this flag multiple
times for multiple variables.
-s, --save_dir PATH Path to a directory to save the requested files.
Extension for the outputs is .nc for geometry and .csv
for coords.
--disable_ssl Pass to disable SSL certification verification.
-h, --help Show this message and exit.
Now, let’s see how we can use PyDaymet as a library.
PyDaymet offers two functions for getting climate data; get_bycoords and get_bygeom.
The arguments of these functions are identical except the first argument where the latter
should be polygon and the former should be a coordinate (a tuple of length two as in (x, y)).
The input geometry or coordinate can be in any valid CRS (defaults to EPSG:4326). The
dates argument can be either a tuple of length two like (start_str, end_str) or a list of
years like [2000, 2005]. It is noted that both functions have a pet flag for computing PET
and a snow flag for separating snow from precipitation using
Martinez and Gupta (2010) method.
Additionally, we can pass time_scale to get daily, monthly or annual summaries. This flag
by default is set to daily.
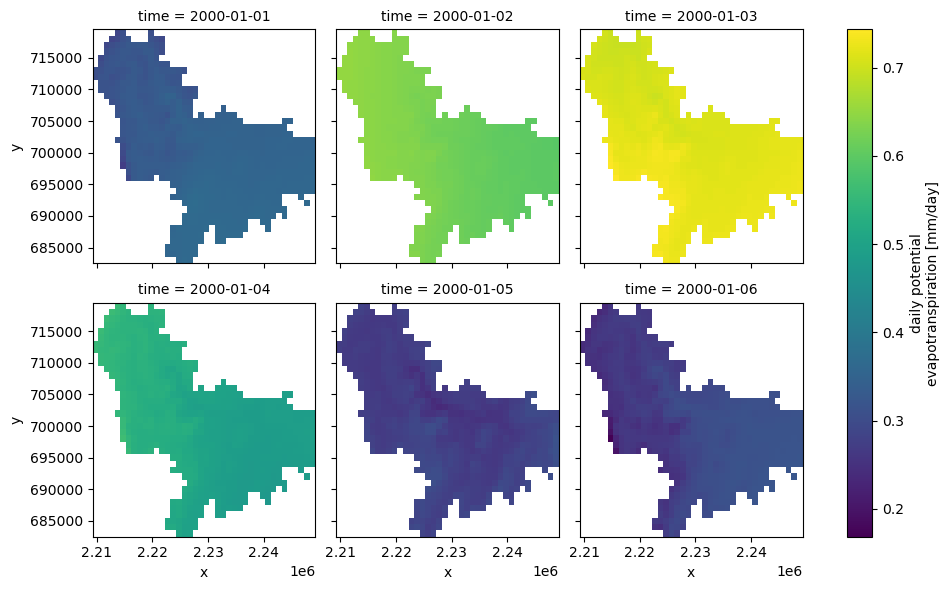
from pynhd import NLDI
import pydaymet as daymet
geometry = NLDI().get_basins("01031500").geometry[0]
var = ["prcp", "tmin"]
dates = ("2000-01-01", "2000-06-30")
daily = daymet.get_bygeom(geometry, dates, variables=var, pet="priestley_taylor", snow=True)
monthly = daymet.get_bygeom(geometry, dates, variables=var, time_scale="monthly")
If the input geometry (or coordinate) is in a CRS other than EPSG:4326, we should pass
it to the functions.
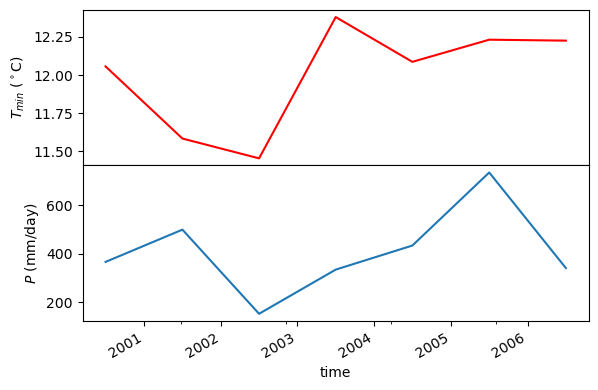
coords = (-1431147.7928, 318483.4618)
crs = 3542
dates = ("2000-01-01", "2006-12-31")
annual = daymet.get_bycoords(coords, dates, variables=var, loc_crs=crs, time_scale="annual")
Additionally, the get_bycoords function accepts a list of coordinates and by setting the
to_xarray flag to True it can return the results as a xarray.Dataset instead of
a pandas.DataFrame:
coords = [(-94.986, 29.973), (-95.478, 30.134)]
idx = ["P1", "P2"]
clm_ds = daymet.get_bycoords(coords, range(2000, 2021), coords_id=idx, to_xarray=True)
Also, we can use the potential_et function to compute PET by passing the daily climate data.
We can either pass a pandas.DataFrame or a xarray.Dataset. Note that, penman_monteith
and priestley_taylor methods have parameters that can be passed via the params argument,
if any value other than the default values are needed. For example, default value of alpha
for priestley_taylor method is 1.26 (humid regions), we can set it to 1.74 (arid regions)
as follows:
pet_hs = daymet.potential_et(daily, methods="priestley_taylor", params={"alpha": 1.74})
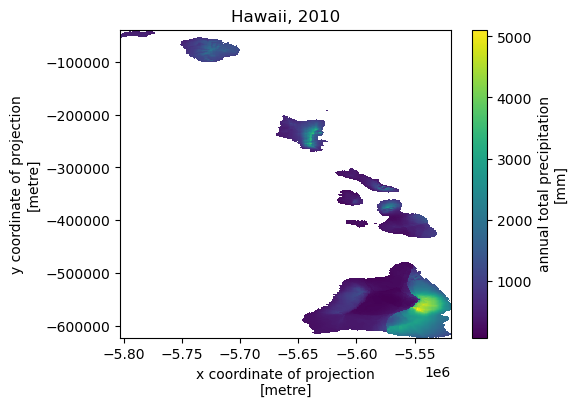
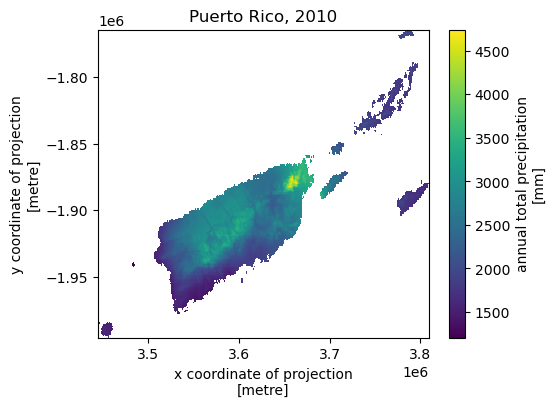
Next, let’s get annual total precipitation for Hawaii and Puerto Rico for 2010.
hi_ext = (-160.3055, 17.9539, -154.7715, 23.5186)
pr_ext = (-67.9927, 16.8443, -64.1195, 19.9381)
hi = daymet.get_bygeom(hi_ext, 2010, variables="prcp", region="hi", time_scale="annual")
pr = daymet.get_bygeom(pr_ext, 2010, variables="prcp", region="pr", time_scale="annual")
Some example plots are shown below: